Retrieving experimental STEM meta data from FEI TIA EMI files
FEI STEM microscopes using TIA to save data in a EMI/SER file pairs. The structure of the SER files is published allowing the image/spectroscopy data to be read using open source third party codes written for python, Matlab, EMD Viewer, ImageJ, and many more.
However, the SER file only contains the pixel size and no other experimental details (meta data) such as the magnification, camera length, accelerating voltage, etc. I have not found a way to retrieve the image/spectroscopy data from the EMI file, but the meta data is human readable as ASCII text. Here is how to retrieve your meta data:
- Open the EMI file in an advanced raw text reader such as Notepad++.
- Note: I do not suggest using the pre-installed Windows apps such as Notepad or Wordpad. Reading image data as text in these programs sometimes makes them crash.
- You will see a lot of random text (like below) where the program interpreted your data as human readable characters:
- PD¢GQH%GHõD?GþIáI{K¸JâLÌM‚OwPzSvTùQ‰TLRœPtPçR¯N
- Search the file for this set of characters:
- <ObjectInfo>
- All text between that set of characters and the ending set </ObjectInfo> is your experimental meta data written in plain text in a structure called XML. To find the values you want just look for “tags” written as <tagname> with the value written next to it.
- For example, 80 kV accelerating voltage will look like:
- <AcceleratingVoltage>80000.0</AcceleratingVoltage>
- For example, 80 kV accelerating voltage will look like:
This is the best way I have found to work with the meta data in EMI files. I think the meta data can be parsed by an XML reader, but I have not attempted to do that properly yet. If someone gets that working then please write back in the comments to let us know how you did it.
Open source DM3 and SER file reading
I work a lot with electron microscopy data files in the scripting languages Matlab and Python. This is a very powerful way to analyze your data, but first you need to get the data read properly. I have posted my own file reading codes in a new Bitbucket repository called openNCEM. The codes are free to use in any way you want. I will post other codes for data analysis using this same repository in the future. Some other NCEM staff and users might add their own code, too.
DM3 files from Gatan’s Digital Micrograph are ubiquitous in the electron microscopy community. Some other programs support this file type, but full support for multidimensional files is not always available. The script called dm3Reader.m will read DM3 files into Matlab as a struct. The struct variable will contain the data and some important metadata from the file tags. Sorry, I have not ported the code to Python yet.
FEI’s TIA software saves data as a proprietary EMI file with accompanying SER files. The EMI files are not readable outside of FEI’s TIA program (yet!), but the SER files can be read. The scripts named serReader.py (for python) and serReader.m (for Matlab) will read the binary data and metadata available in the SER file. Supported experiment types are images, image stacks, spectra, spectral line scans and spectral maps. The output of the script is a python dictionary or Matlab struct.
Let me know if you have trouble loading any files. The codes are not extensively tested with data from other institutions. Try them out, and leave any suggestions in the comments here.
Molecular Foundry Proposal Submission
The National Center for Electron Microscopy (NCEM) will merge with The Molecular Foundry (MF) on October 1st. Staff members of the two centers have worked together for almost a year to ensure that the merger process goes smoothly.
Coincidentally, the next deadline for Molecular Foundry (and NCEM) proposals is also on October 1st. Please visit the new proposal submission website to create an account and start the process. Feel free to contact me with any questions about your proposed project or the new submission process.
Electron Microscopy Data Files EMI / SER (update)
Electron Microscopy Data Files EMI / SER (update)
Some time ago I posted about a link to FEI’s ES Vision / TIA software available on their website, which eventually stopped working. It is working again, and the available download is named ES Vision (originally built by Emispec and bought by FEI). This software is used on Tecnai microscopes (not Titans). The available version is 3.0 (different than indicated in the link text), and is much older compared to the most recent TIA 4.0 running on current Titan microscopes. ES Vision can still open some EMI / SER files, but I am not sure of compatibility.
I highly suggest that you contact your FEI service representative to receive an off-line copy of the newest TIA 4.0 (or higher) installation.
To install ES Vision:
Download the ES Vision Off-line full installation. Run the EXE file, which extracts numerous files to C:\temp\ESVTemp\ by default. Navigate to that directory and then open the ESVision directory (ignore everything else). Run SETUP.exe to install the software. The password is LastChanceGulch (case sensitive). I am assuming that FEI wants you to install this software, so I am posting the password. Note: I have not been able to load TIA or ES Vision onto Windows 8 computers. (Second note: I have been notified that the password no longer works as of March, 2017. It may still work on older versions, so Ill keep this information here.)
SER files are relatively easy to read without TIA or ES Vision and contain the binary data and pixel size information. However, the EMI file contains extra information, which is often very useful.
Electron tomography short course
Electron tomography short course
I am excited to announce that NCEM / MF are holding a 3-day short-course on electron tomography from January 16 – 18, 2013.
Deadline to enroll: Monday, December 17, 2012
Click the link above for more information.
3D Tomography of polymers in Micron
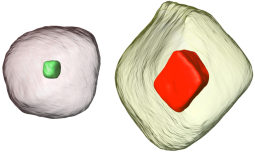
An article recently published in the journal Micron incorporates 3D electron tomography I did in collaboration with Frances Allen at NCEM. We used tomography to investigate very different morphologies of block copolymer systems cast from different solvents. Importantly, the films were spun onto substrates incorporating holes overwhich the films grew, which provides a symetric boundary condition of two free surfaces. The image above shows two orthogonal slices through a polymer film with unexpected 3D layered morphology with a semi-solid phase sandwiched between a honeycomb-like phase. 3D analysis is key in determining the full 3D structure of such complex polymer systems.
Visit by John Murray of Senator Diane Feinstein’s SF office
I gave a tour of NCEM to John Murray of Diane Feinstein’s office last week. It is great to see interest from senator’s regarding scientific instruments and progress at LBNL. I enjoyed chatting with Mr. Murray and showing him our lab.
New publication on TEAM I
A new article detailing the TEAM I microscope was just published that I wrote with the help of colleagues at the National Center for Electron Microscopy. Included in the write-up are details about the stability and performance of the correctors and the TEAM Stage. There are also some results showing the resolution of the microscope at low accelerating voltages. The article was published as part of a special issue on aberration corrected electron microscopy.
Electron Microscopy Data Files (DM3 and EMI / SER)
Most FEI electron microscopes acquire data with either Gatan Digitalmicrograph (for TEM) and ES Visision TIA (for STEM). Gatan files have a DM3 file extension and TIA files have EMI/SER file extensions. FEI and Gatan previously did not offer any free (or even cheap) way to get access to you data easily. That has finally changed, and now you can read DM3 and EMI/SER files on your own computer with offline software versions for free.
FEI released ES Vision version 4.0.173, which you can install on a windows computer. Be sure to run the program as an administrator (especially in Windows 7) or you will run into some error messages.
Gatan released a basic version of Digitalmicrograph (DM) as well. Their website states: “you can install and use DM in an offline context (no hardware attached) at no charge.” You first need to request an offline demo license at the Digitalmicrograph Offline Request webpage. After filling out the form, solve hte captcha and press submit at the bottom of the page. Click on the demo license (32-bit or 64-bit) which you require for your operating system. Extract the license ZIP file to a folder, read the Read-me.PDF file created and install the license with LicenseInstaller.exe. Next, download the latest 2.X version of the (Gatan Microscopy Suite (GMS) from Gatan’s FTP site and install the software.
You can finally work with your data away from the microscope without spending a lot of money for a software license. Also, I have written a few Matlab scripts to import images, spectra, data cubes and all associated tags. Just contact Peter Ercius or you can download a FEI TIA SER reader for many different programs at the openNCEM Bitbucket repository.
TEM electron tomography of ordered nanorods at NCEM
TEM electron tomography of ordered nanorods at NCEM
A recent paper I was involved with was published in Nano Letters and featured on the LBL website. We used TEM electron tomography to reconstruct nanorods in a block copolymer matrix. The polymer self assembles into arrays which makes it easier to produce ordered structures of nanorods. The technique could be used to improve solar cells, sensors and mechanical materials properties. The original tomography results were published in Nano Letters.